Editor: Nina
This study reveals that exosomes derived from CD34+ hematopoietic stem/progenitor cells interact with E-selectin in a calcium-dependent manner, facilitating their targeted migration to specific organs, thereby highlighting their potential role in leukemia progression and therapeutic applications.
Key Preview
Research Question
The study titled “CD34+ HSPCs-derived exosomes contain dynamic cargo and promote their migration through functional binding with the homing receptor E-selectin” aims to investigate how exosomes derived from hematopoietic stem/progenitor cells (HSPCs) can influence cell migration and homing by interacting with the E-selectin receptor.
Research Design and Strategy
The researchers employed biochemical and imaging techniques to analyze the cargo of exosomes isolated from both leukemic and healthy CD34+ HSPCs and assess their migration capabilities.
Method
Exosomes were isolated using a serial centrifugation approach, followed by proteomic analysis to identify their cargo. Binding assays and in vivo experiments were conducted to determine the role of E-selectin in exosome migration.
Key Results
The study found that CD34+ HSPC-derived exosomes express E-selectin ligands and can bind to E-selectin in a calcium-dependent manner. When injected into mice, these exosomes were found to target organs such as the spleen and spine, indicating a potential role in leukemic cell engraftment.
Significance of the Research
These findings suggest that exosomes derived from HSPCs not only serve as carriers of molecular signals but also play an active role in cell migration and homing. This research could open new avenues for targeted therapies in leukemia treatment and understanding metastasis.
Introduction
Exosomes are small extracellular vesicles that facilitate intercellular communication by transporting proteins, lipids, RNA, and other molecules. Their role in cancer biology, particularly in leukemia, has garnered attention, as these exosomes can alter the tumor microenvironment and promote disease progression. Previous studies have suggested that leukemic cells use exosomes to prepare distant sites for engraftment, but the mechanisms underlying the initial steps of this migration process remained poorly understood. This study aims to elucidate how exosomes derived from CD34+ HSPCs bind to the homing receptor E-selectin, which is essential for their migration.
Research Team and Objective
The research was conducted by a team comprising Ioannis Isaioglou, Mansour M. Aldehaiman, Yanyan Li, and others from the King Abdullah University of Science and Technology (KAUST). Published in Frontiers in Cell and Developmental Biology, the study investigates the dynamic cargo of CD34+ HSPC-derived exosomes and their functional binding to E-selectin, focusing on implications for cell migration and potential therapeutic applications in leukemia.
Experimental Process
1. Cell Culture
Key Steps:
- The KG-1a cell line, a CD34+ acute myeloid leukemia (AML) cell line, and K562 cells were obtained from the American Type Culture Collection (ATCC).
- KG-1a cells were cultured in Roswell Park Memorial Institute (RPMI 1640) media, while K562 cells were cultured in Iscove’s Modified Dulbecco’s Medium (IMDM), both supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin.
- Cells were maintained at a density of 0.8 × 10^6 cells/ml in a humidified incubator at 37°C with 5% CO2.
Results and Key Data:
- The successful culture of KG-1a and K562 cell lines ensured that a sufficient quantity of viable cells was available for subsequent experiments.
Significance of the Result:
- Using these well-established cell lines provided a reliable model for studying exosome production and functionality in the context of leukemia.
Key Innovations:
- This approach overcomes the limitations of traditional cell lines by utilizing primary human CD34+ cells from healthy and leukemic donors, enhancing translational relevance.
2. Exosome Isolation
Key Steps:
- Exosomes were isolated using a serial centrifugation protocol. Initially, cell culture media was centrifuged at 300 x g for 10 minutes to remove cells, followed by a 2000 x g spin for 30 minutes, and a final ultracentrifugation at 100,000 x g for 120 minutes to pellet the exosomes.
- The exosome pellet was washed and resuspended in 150 μl of PBS for downstream assays.
Results and Key Data:
- Isolation yielded exosomes confirmed by dynamic light scattering (DLS), scanning electron microscopy (SEM), and Western blot analysis, showing enriched exosomal markers such as CD63 and CD81.
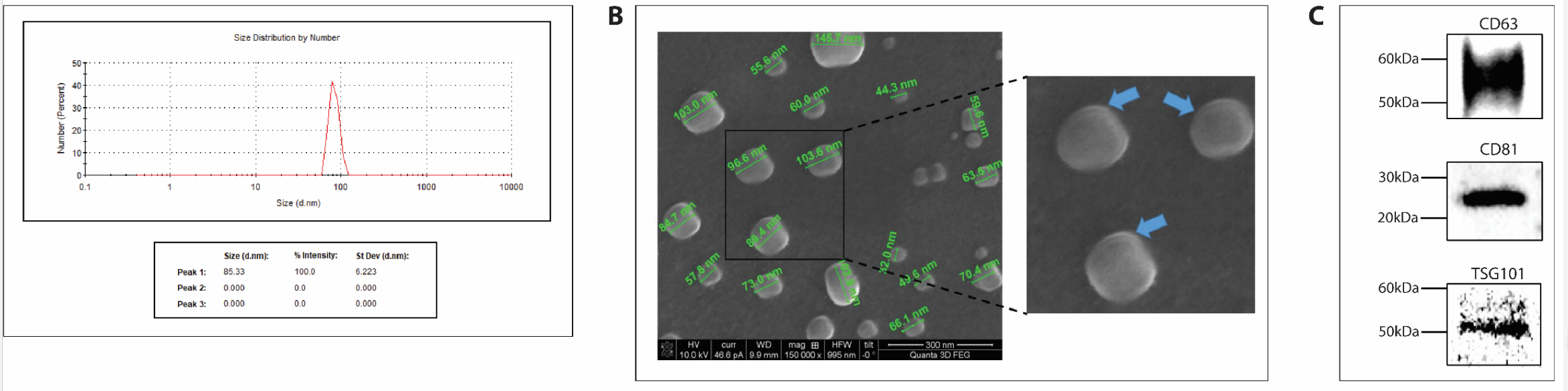
Figure 1. Characterization and quality control of cell derived exosome preparations.
Significance of the Result:
- This method produced high-purity exosomes suitable for detailed characterization and functional assays.
Key Innovations:
- The use of ultracentrifugation optimizes exosome yield and purity compared to conventional precipitation methods, enhancing the quality of the isolated vesicles for subsequent applications.
3. Proteomic Analysis
Key Steps:
- Isolated exosomes underwent mass spectrometry for proteomic profiling. Proteins were separated using SDS-PAGE, and gel bands were excised for in-gel digestion with trypsin.
- The resulting peptides were analyzed using a Q-Exactive HF mass spectrometer.
Results and Key Data:
- Approximately 2000 proteins were identified in the exosomal proteome, including adhesion-related proteins such as CD34 and CD44, as well as signaling molecules involved in migration.
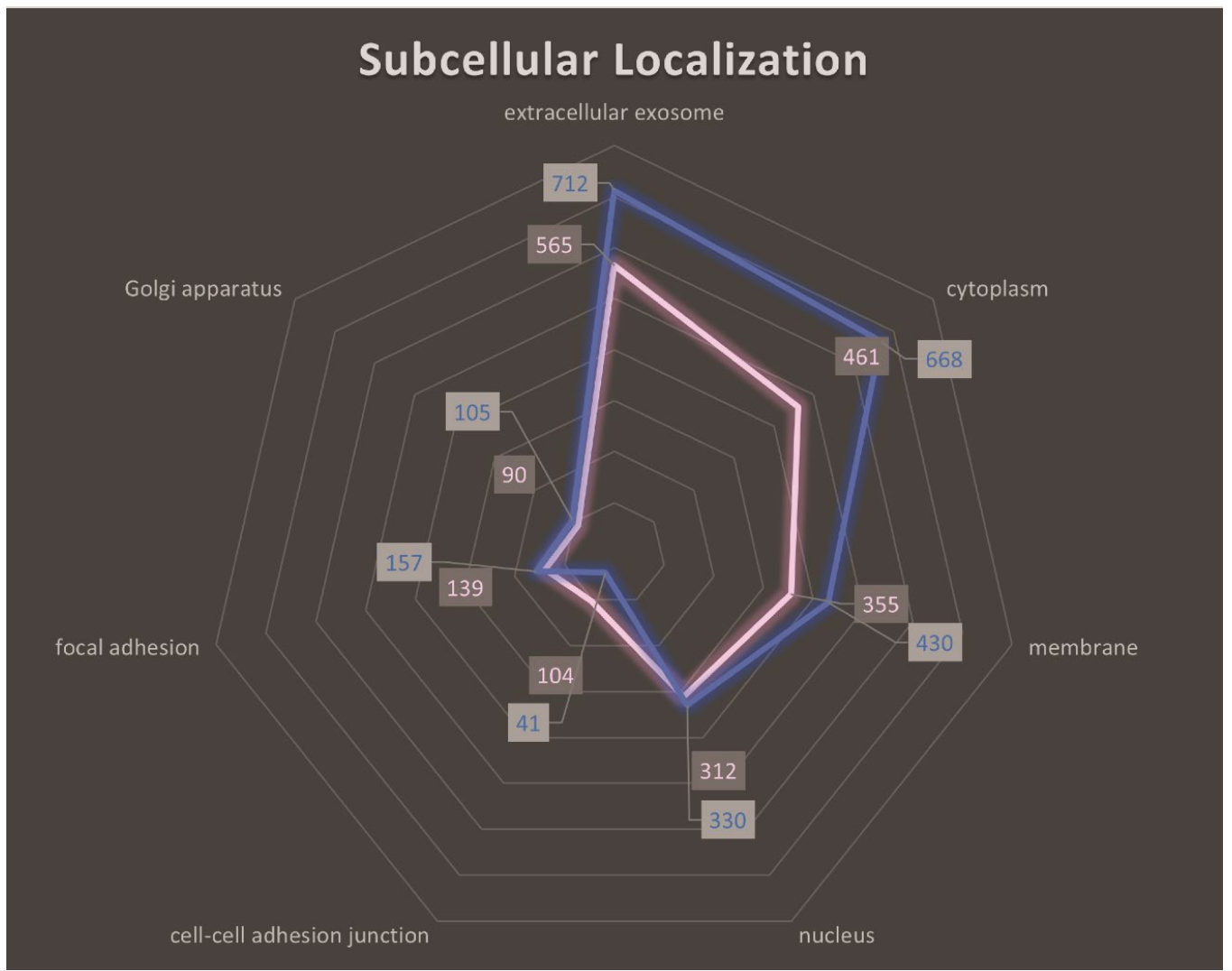
Figure 2. Subcellular localization analysis of HSPCs-derived exosomal proteins. Radar diagram illustrating the top seven subcellular localization candidates of the exosomal proteome (KG1apink, healthy HSPCs-blue). The closer to the edge of the diagram, the more proteins are related to the particular subcellular component referenced. The “extracellular exosome” category generated the highest hit confirming the successful isolation of exosomes.
Significance of the Result:
- This comprehensive proteomic analysis revealed that exosomes carry a cargo relevant to cell adhesion and migration, supporting the hypothesis that exosomes contribute to leukemic cell homing.
Key Innovations:
- The integration of mass spectrometry allows for a detailed understanding of exosomal cargo, facilitating the identification of potential therapeutic targets, unlike traditional methods that often provide limited insights into exosome composition.
4. Binding Assays
Key Steps:
- Binding assays were conducted using recombinant E-selectin (rE-selectin) to test the interaction with exosomal ligands. Exosomes were incubated with rE-selectin in the presence of calcium or EDTA to assess calcium dependency.
- Protein immunoprecipitation was performed with rE-selectin-IgG to isolate bound proteins for Western blot analysis.
Results and Key Data:
- The results confirmed calcium-dependent binding of exosomes to E-selectin, with specific ligands (CD34, CD44, PSGL-1) detected in the bound fraction.
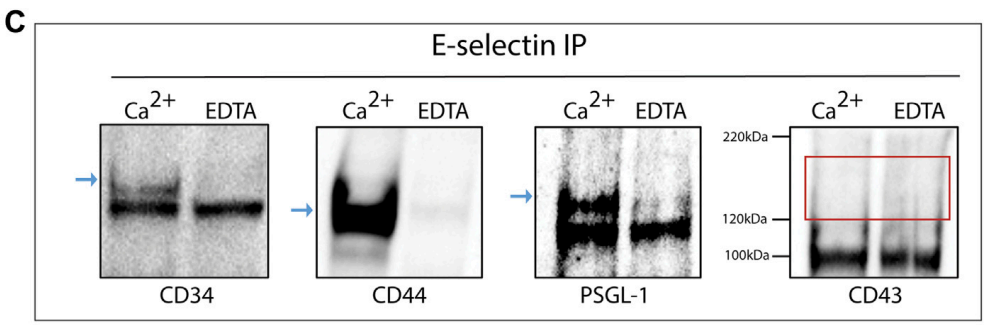
Figure 3. rE-selectin-IgG was used to immunoprecipitate (IP) proteins from lysates of KG1a-derived exosomes either in the presence of Ca2+ (2 mM) or EDTA (20 mM). Western blot analysis revealed the presence of CD34, CD44, and PSGL-1 (blue arrows) in samples where Ca2+ was added but not in samples containing EDTA confirming the selective, calcium-dependent binding of E-selectin to its ligands. In western blots for CD43, no band was observed at the expected MW (see red rectangle).
Significance of the Result:
- Establishing the binding of exosomes to E-selectin demonstrates their potential role in facilitating leukemic cell migration to specific tissues, providing insights into hematological malignancies.
Key Innovations:
- This experimental design allows for real-time analysis of exosomal interactions in a physiological context, vastly improving upon static in vitro adhesion assays that fail to replicate in vivo dynamics.
5. In Vivo Studies
Key Steps:
- NSG mice were divided into three groups: a control group receiving PBS, a group receiving exosomes, and a group pretreated with anti-E-selectin antibodies followed by exosome injection.
- Biodistribution of fluorescently labeled exosomes was tracked using IVIS imaging at multiple time points (2, 6, 12, 24, and 48 hours post-injection).
Results and Key Data:
- Exosomes showed significant accumulation in the spleen and spine of mice, indicating targeted delivery facilitated by E-selectin interaction. The accumulation was markedly reduced in the presence of the blocking antibody.
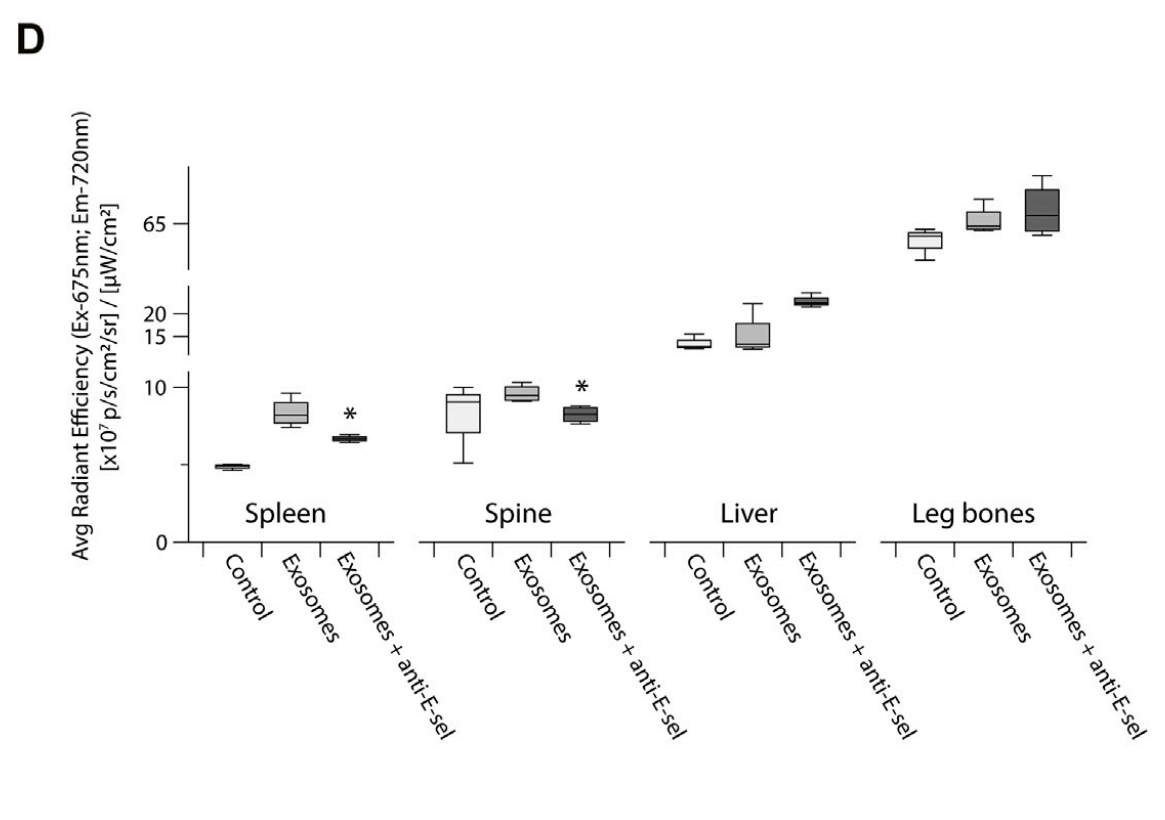
Figure 4. Quantitative analysis of the fluorescence intensity of the leg bones, spine, spleen and liver are shown the exosomes accumulation in different organs and were statistically analyzed by student’s t -test (p-value (*) <0.05, Exosomes group vs. Exosomes + anti-E-sel group).
Significance of the Result:
- These findings highlight the organotropic behavior of exosomes and their dependency on E-selectin for targeted delivery, which is essential for therapeutic applications in leukemia.
Key Innovations:
- Utilizing E-selectin as a targeting mechanism for exosomes represents a novel strategy for drug delivery systems, enhancing specificity and minimizing off-target effects compared to traditional nanoparticle delivery methods.
Conclusion
The research highlights that CD34+ HSPC-derived exosomes possess the ability to interact with E-selectin, facilitating migration to specific organs. This interaction is crucial for understanding the mechanisms of leukemic cell homing and could guide the development of targeted therapies. However, the study acknowledges limitations, such as the complexity of in vivo environments and the need for further exploration of the functional impacts of altered exosomal cargo. Future research could focus on broadening the understanding of exosomal modifications and their implications in cancer metastasis, enhancing therapeutic interventions based on these findings.
Reference
Isaioglou, Ioannis, et al. “CD34+ HSPCs-derived exosomes contain dynamic cargo and promote their migration through functional binding with the homing receptor E-selectin.” Frontiers in Cell and Developmental Biology 11 (2023): 1149912.
