Editor: Nina
A study by King Abdullah University of Science and Technology explores how exosomes, tiny cell-derived vesicles, can be engineered to deliver targeted therapies to sites of Acute Myeloid Leukemia (AML) by utilizing E-selectin binding for improved treatment efficacy.
Key Preview
Research Question
The research investigates how exosomes, derived from CD34+ hematopoietic stem/progenitor cells, can be engineered for targeted delivery in Acute Myeloid Leukemia (AML) by utilizing their interaction with E-selectin.
Research Design and Strategy
The researchers designed a series of experiments to isolate exosomes from both healthy and AML-derived cells, analyzing their interaction with E-selectin receptors to determine their suitability for targeted drug delivery.
Method
Exosomes were isolated using centrifugation, and their surface proteins were analyzed via proteomics. The exosome-E-selectin interaction was studied through various biochemical techniques, including immunoprecipitation, Western blot, and flow assays.
Key Results
The study found that exosomes from AML cells carry E-selectin ligands, enabling them to bind and migrate to sites of leukemia, such as the spleen and spine. Moreover, modifying the exosome’s surface glycosylation with recombinant fucosyltransferase enhanced their binding affinity for E-selectin.
Significance of the Research
This innovative exosome-based drug delivery system could overcome limitations of current therapies like chemotherapy by providing more precise targeting, minimizing side effects, and potentially improving AML treatment outcomes.
Introduction
Acute Myeloid Leukemia (AML) is a form of cancer that affects the blood and bone marrow, leading to the rapid growth of abnormal white blood cells. Traditional treatments, including chemotherapy, often come with significant drawbacks such as poor targeting of cancer cells, widespread side effects, and limited efficacy.
A promising alternative to these methods is the use of exosome-based drug delivery systems. Exosomes, which are tiny vesicles secreted by cells, can carry therapeutic molecules and, due to their natural ability to interact with specific cell receptors, offer a more targeted approach for drug delivery. This research proposes the use of exosomes to directly target AML cells through their binding to E-selectin, a receptor commonly expressed on endothelial cells in the bone marrow and sites of leukemia.
Research Team and Aim
The research team is led by Dr. Jasmeen S. Merzaban from the Bioscience Program at King Abdullah University of Science and Technology (KAUST), Saudi Arabia. The study, titled “CD34+ HSPCs-derived exosomes contain dynamic cargo and promote their migration through functional binding with the homing receptor E-selectin”, was published in Frontiers in Cell and Developmental Biology in April 2023.
According to Dr. Merzaban, the goal of the research was to explore the potential of exosomes, derived from both healthy and AML-derived hematopoietic stem/progenitor cells, as vehicles for targeted delivery of therapeutic agents to sites of leukemia, improving treatment precision and reducing harmful side effects associated with conventional therapies.
Experimental Process
Primary Technique: Exosome Isolation and Functional Analysis
The research employed a primary technique of exosome isolation via a series of centrifugation steps. This method was fundamental to extracting exosomes from both healthy and AML-derived hematopoietic stem/progenitor cells (HSPCs) for subsequent functional analysis. Exosome isolation began with culturing cells in serum-free media, followed by a series of centrifugation steps to remove larger particles. The final exosome fraction was obtained by ultracentrifugation, ensuring a highly pure exosome population for further investigation.
Key Steps
1.Exosome Isolation: The first step involved culturing cells (e.g., KG1a, a leukemia cell line) in media without fetal bovine serum (FBS) for 48 hours to induce exosome release. The culture supernatant was then subjected to several centrifugation steps to remove cellular debris and larger vesicles.
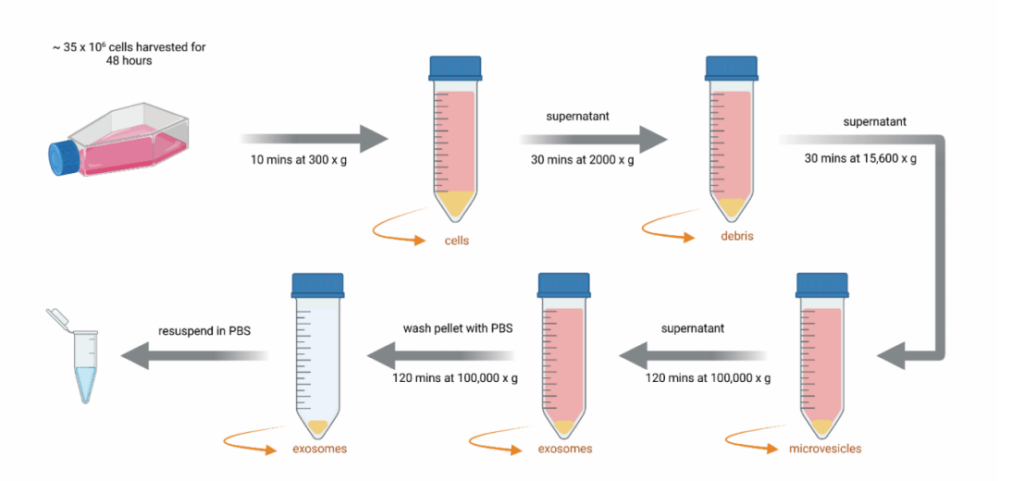
Figure 1. Exosome isolation process. Cartoon illustrating the isolation protocol of exosomes derived from cell types used throughout the manuscript. Following a starvation of the cells for 48 hours, serial centrifugation steps took place to remove larger particles from the media. By the last ultracentrifugation step, exosomes were pelleted and finally resuspended in PBS buffer.
2. Exosome Characterization: After isolation, the exosomes were characterized using techniques like Dynamic Light Scattering (DLS) and Scanning Electron Microscopy (SEM) to confirm their size and morphology. The key goal was to ensure that the isolated particles were within the typical exosome size range (40-160 nm) and were free from contaminating proteins.
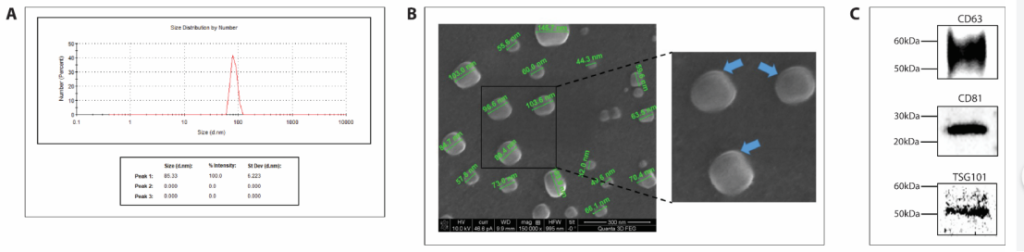
Figure 2. Characterization and quality control of cell derived exosome preparations.
3.Proteomic Analysis: The next step involved mass spectrometry analysis to determine the protein content of the exosomes. This revealed several proteins related to adhesion and migration, such as integrins, selectin ligands (e.g., CD34, CD44), and chemokines.
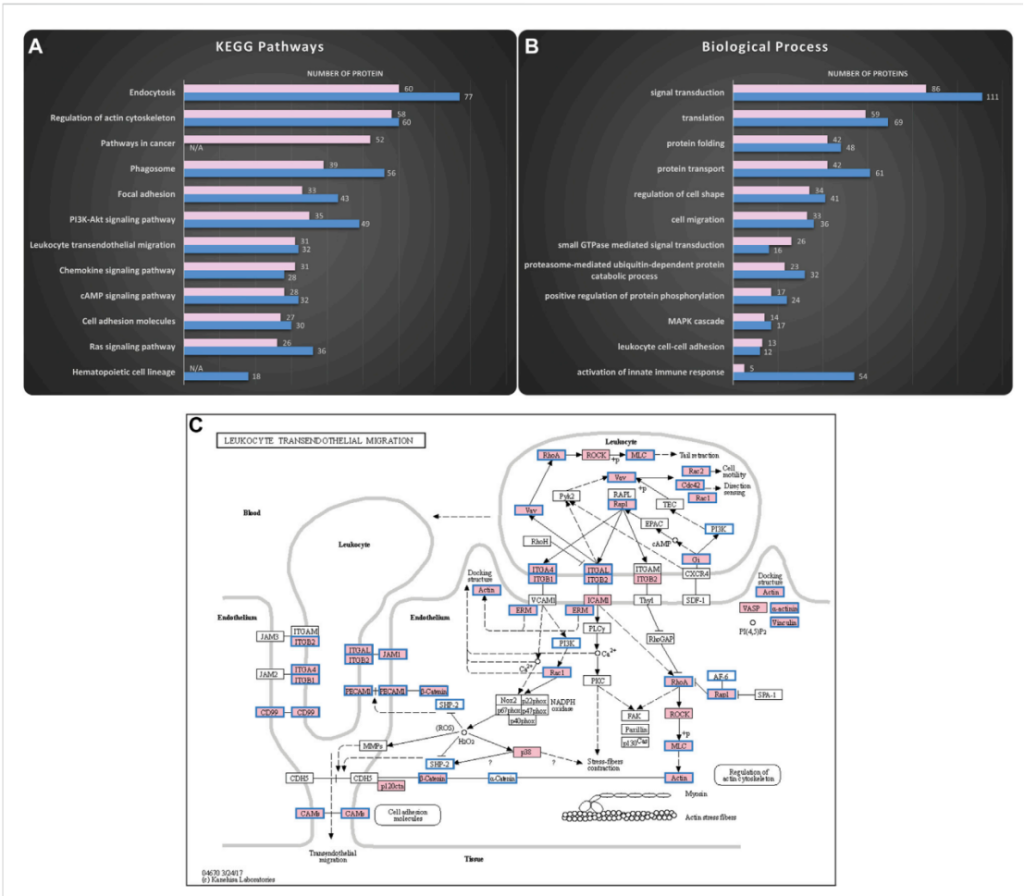
Figure 3. Mass spectrometry analysis reveals HSPCs-derived exosomes are enriched in proteins involved in adhesion and migration. Proteomics analysis of the cargo was performed to uncover putative functions of the isolated exosomes. The KEGG pathways and the Biological Processes from Gene Ontology analysis that were enriched in the KG1a (pink) and healthy HSPCs (blue)-derived exosomes were identified using the DAVID online tool.
Data Collection and Analysis
For this study, Western blotting was used extensively to analyze exosomal proteins. The exosomes were lysed, and the protein content was probed for specific markers, including E-selectin ligands. The binding of exosomes to recombinant E-selectin-IgG was assessed by immunoprecipitation, which allowed the researchers to confirm the interaction between exosomes and E-selectin. Furthermore, fluorescence microscopy and live imaging were used in in vivo experiments to track the migration of labeled exosomes in mice, offering real-time insights into their biodistribution. The collected data were quantitatively analyzed using standard statistical tests to evaluate differences between experimental conditions.
Novel Aspects and Advantages
One of the most innovative aspects of this study is the modification of exosomes to enhance their binding to E-selectin through dynamic glycosylation. The researchers employed recombinant fucosyltransferase to modify the surface of exosomes, enabling them to express sialyl-Lewis X (sLex) epitopes, which are necessary for binding to E-selectin. This modification significantly increased the exosome’s affinity for E-selectin and allowed for more precise targeting to sites associated with AML, such as the spleen and spine.
Additionally, the study’s use of exosomes from both healthy and AML-derived cells is novel in the context of targeted drug delivery. Traditional nano-delivery systems often suffer from poor specificity, leading to off-target effects. In contrast, the exosome-based approach harnesses the natural targeting abilities of exosomes, offering a more refined system for delivering therapeutic agents directly to cancerous tissues. This method not only improves the targeting precision but also minimizes the side effects typically associated with conventional treatments like chemotherapy
Conclusion
This research successfully developed an exosome-based delivery system that can target AML cells more precisely than traditional chemotherapy methods. The study demonstrated that exosomes, through their natural ability to bind E-selectin, can effectively deliver therapeutic cargo to leukemia sites such as the spleen and spine. The findings highlight the potential of this system in improving the treatment outcomes for AML by offering more targeted therapies, which could significantly reduce the side effects often caused by conventional treatments.
This exosome-based approach also suggests that dynamic modifications to the exosomal cargo, such as altering glycosylation patterns, could provide further improvements in targeting and therapeutic efficacy, potentially opening the door to new methods for treating various cancers and other diseases.
Reference
Isaioglou, Ioannis, et al. “CD34+ HSPCs-derived exosomes contain dynamic cargo and promote their migration through functional binding with the homing receptor E-selectin.” Frontiers in Cell and Developmental Biology 11 (2023): 1149912.
