A study demonstrates that GalNAc-Lipid nanoparticles can achieve efficient liver-targeted delivery of CRISPR base editing therapy, offering a potential option for treating familial hypercholesterolemia and other liver diseases.
Background
Challenges in Delivering Genetic Therapies to the Liver
Familial hypercholesterolemia (FH) is a rare, inherited disorder caused by mutations in the low-density lipoprotein receptor (LDLR) gene, leading to elevated LDL cholesterol levels and increased cardiovascular risk. For patients with homozygous familial hypercholesterolemia (HoFH), current therapies are often ineffective, as they depend on residual LDLR activity, which is significantly impaired or absent in this population.
Lipid nanoparticles (LNPs) have emerged as a preferred strategy for delivering genetic therapies to the liver. However, most LNPs rely on LDLR-mediated endocytosis for uptake into hepatocytes, limiting their efficacy in patients with LDLR deficiencies. CRISPR-based therapies represent a promising alternative, allowing for precise, one-time genetic editing. Nevertheless, efficient delivery to hepatocytes independent of LDLR activity remains a significant obstacle.
Innovation with GalNAc-Lipid Nanoparticles
This study addresses this challenge by employing GalNAc-Lipid nanoparticles (GalNAc-LNPs), which leverage asialoglycoprotein receptor (ASGPR)-mediated uptake. ASGPR is highly expressed on hepatocytes and offers an alternative pathway for liver-targeted delivery. Unlike LDLR-dependent methods, ASGPR targeting ensures delivery precision and minimizes off-target effects.
Research Aim & Objectives
Aim and Focus
The primary aim of the research was to develop an effective LDLR-independent delivery system for CRISPR base editing therapy, targeting the ANGPTL3 gene in hepatocytes. ANGPTL3 is a regulator of lipid metabolism, and its inactivation has been shown to reduce plasma lipid levels, including LDL cholesterol.
Research Team and Publication Details
The study was conducted by researchers from Verve Therapeutics and the Perelman School of Medicine at the University of Pennsylvania. The findings were published in Nature Communications in 2023.

Research Methods & Results
Experimental Process: Overview
- Structure-guided optimization of GalNAc-LNPs.
- In vivo screening of liver-targeted editing in LDLR-deficient mice.
- Validation in wild-type and LDLR-deficient non-human primates (NHPs).
- Safety and durability assessments over several months.
Key Experiments
- Structure-Guided Design of GalNAc-LNPs:
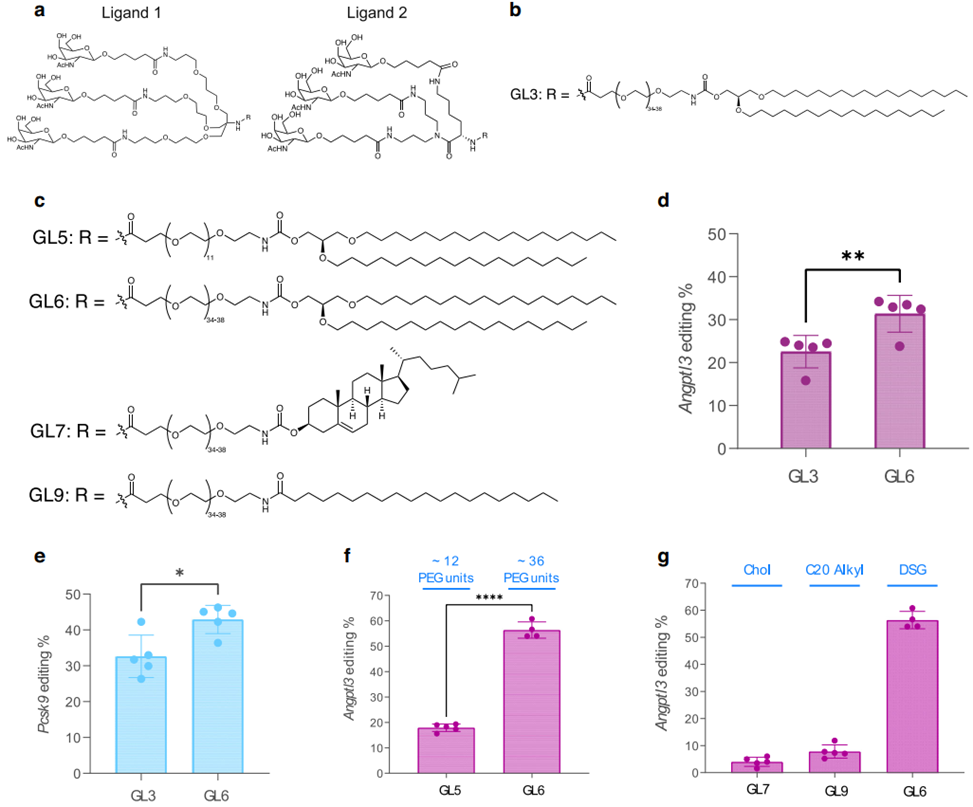
The research team created two ligand designs—Design 1 (TRIS scaffold) and Design 2 (lysine scaffold). Ligand Design 2, which featured a simpler and more scalable synthesis process, was used to develop the optimized GalNAc-Lipid GL6. To test the efficacy, GL6 and GL3 (based on Design 1) were formulated into LNPs and injected into LDLR-deficient mice. GL6 achieved significantly higher liver editing of the ANGPTL3 gene (31%) compared to GL3 (23%, p = 0.0086). This demonstrated the superiority of ligand Design 2 in improving ASGPR-mediated uptake. - Optimization of PEG Spacers and Lipid Anchors:
To improve delivery efficiency, the researchers tested different PEG spacer lengths (12-unit vs. 36-unit) and lipid anchors (DSG, cholesterol, and arachidoyl). In LDLR-deficient mice, LNPs with the longer 36-unit PEG spacer and DSG lipid anchor achieved liver editing rates of 56%, compared to 18% for the shorter PEG spacer. The longer spacer improved receptor binding, while the DSG anchor enhanced nanoparticle stability, making this combination the optimal configuration for GalNAc-LNPs.
- Validation in LDLR-Deficient NHPs:
The optimized GalNAc-LNPs were evaluated in LDLR-deficient NHPs to model HoFH. In these primates, GalNAc-LNPs delivered CRISPR base editors targeting the ANGPTL3 gene, resulting in a 61% liver editing rate and an 89% reduction in plasma ANGPTL3 protein. This reduction was accompanied by a stable 35% decrease in LDL cholesterol over three months, underscoring the platform’s therapeutic potential for patients with LDLR deficiencies.
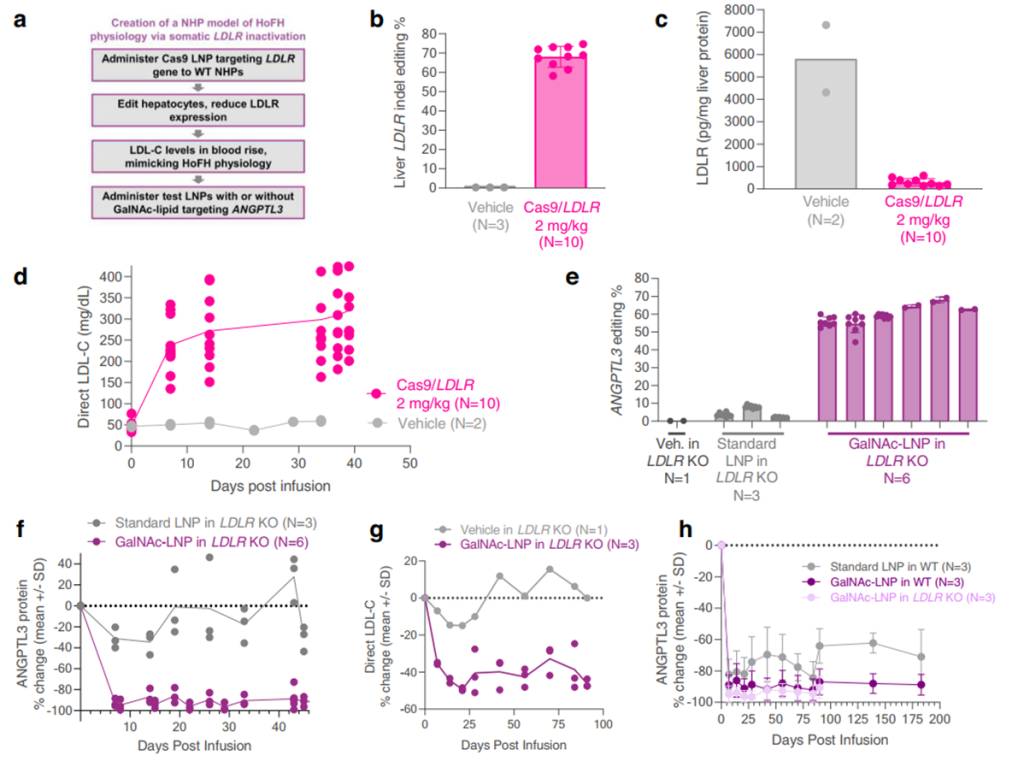
- Durability and Safety Testing:
The durability of GalNAc-LNPs was assessed in wild-type NHPs over six months. Liver editing rates of 64% and reductions in ANGPTL3 protein levels of 90% were observed after a single dose. Transient increases in liver enzymes (ALT and AST) and inflammatory cytokines normalized within two weeks, confirming the platform’s tolerability and safety for repeated use.
Key Technical Points and Innovations
- Multivalent GalNAc Ligands: Enabled efficient ASGPR-mediated uptake into hepatocytes.
- PEG Spacers and Lipid Anchors: The 36-unit PEG spacer and DSG lipid anchor enhanced stability and delivery efficiency.
- LDLR Independence: Comparable editing efficiency across wild-type and LDLR-deficient models demonstrated the system’s versatility.
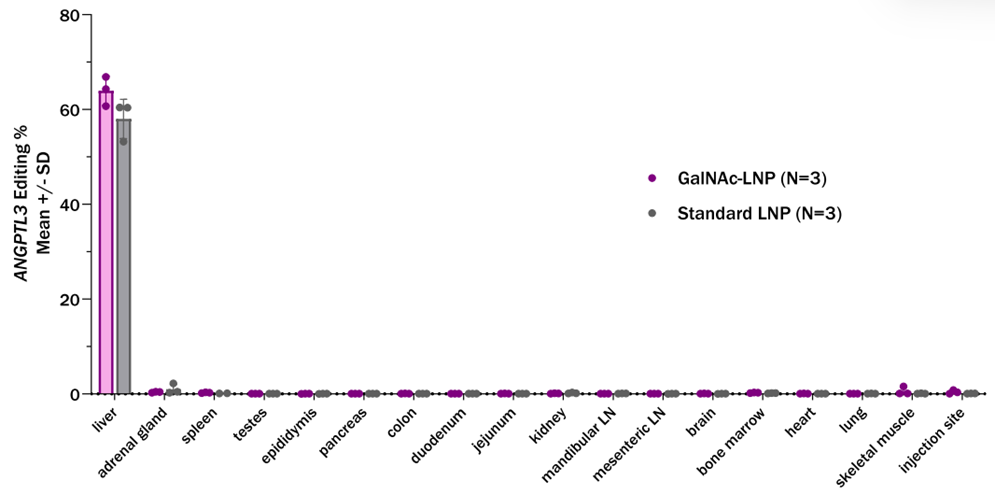
- Target Specificity: Minimal off-target editing (<2%) was observed in non-hepatic tissues, supporting the precision of the delivery system.
Summary
This study successfully developed and validated GalNAc-LNP technology for LDLR-independent hepatic delivery of CRISPR base editing therapy. Key findings include:
- Efficient Liver Editing: GalNAc-LNPs achieved up to 61% gene editing in LDLR-deficient primates, with corresponding reductions in ANGPTL3 protein levels (up to 89%).
- Long-Term Effects: A single dose resulted in sustained therapeutic effects, with significant reductions in plasma LDL cholesterol (~35%) and protein levels lasting up to six months.
- Safety and Specificity: The delivery system exhibited high specificity for liver tissue and a favorable safety profile, with only transient increases in liver enzymes and cytokines.
The innovation of the GalNAc-LNP platform lies in its ability to bypass LDLR-dependent pathways, making it a viable therapeutic option for HoFH patients. Additionally, the platform’s versatility and precision could enable its application to other liver-targeted genetic therapies.
While these findings represent a significant advance, further studies are needed to evaluate potential off-target effects and explore re-dosing strategies to maximize clinical utility. This work provides a foundation for the continued development of safe, efficient, and targeted delivery systems for genome-editing therapies.
Reference:
Kasiewicz, Lisa N., et al. “GalNAc-Lipid nanoparticles enable non-LDLR dependent hepatic delivery of a CRISPR base editing therapy.” Nature Communications 14.1 (2023): 2776.
