This study presents a computationally designed antigen, T2_17, which elicits broad humoral immune responses against SARS-CoV-2 and related sarbecoviruses, demonstrating potential for effective vaccine development to combat zoonotic spillovers and emerging variants.
Key Preview
- Research Question: The study investigates how to develop a vaccine that elicits broad humoral immune responses against SARS-CoV-2 and related sarbecoviruses, which pose a threat of zoonotic spillovers.
- Research Design and Strategy: The researchers employed a viral-genome-informed computational method to select immune-optimized and structurally engineered antigens based on the receptor binding domain (RBD) of the spike protein of sarbecoviruses.
- Method: The study utilized in vivo immunization in animal models (mice, rabbits, and guinea pigs) to evaluate the immunogenicity and protective efficacy of the designed antigens. The optimized antigen was delivered through DNA immunogen or a modified vaccinia virus Ankara vector.
- Key Results: The optimized antigen induced significant cross-reactive antibody responses against multiple strains, including SARS-CoV-1, SARS-CoV-2, WIV16, and RaTG13. Specifically, mice immunized with the T2_17 antigen demonstrated effective neutralization against the Delta variant of SARS-CoV-2.
- Significance of the Research: This research offers a promising strategy for developing vaccines that provide broader protection against emerging coronaviruses, potentially counteracting the threat of zoonotic spillovers and variants of concern.
Introduction
The ongoing COVID-19 pandemic has underscored the urgent need for vaccines capable of offering broader protection against coronaviruses, particularly those that can spill over from animals to humans. Historically, two significant zoonotic events have been linked to sarbecoviruses: the outbreak of severe acute respiratory syndrome (SARS) in 2002 and the current pandemic caused by SARS-CoV-2. As these viruses continue to evolve, the emergence of new variants raises concerns about vaccine efficacy and the potential for future outbreaks.
In light of this, the research team sought to create a vaccine that could elicit robust immune responses across different strains of sarbecoviruses. By leveraging computational design, they aimed to optimize a single antigen based on the receptor binding domain (RBD) of the spike protein, thereby enhancing the likelihood of cross-protection against existing and emerging variants.
Research Team and Objective
The study was conducted by an international team of researchers, including Sneha Vishwanath, George William Carnell, and Jonathan Luke Heeney, among others, affiliated with the University of Cambridge and other institutes. The research was published in Nature Biomedical Engineering. The primary objective was to develop a computationally designed antigen capable of eliciting broad humoral responses against SARS-CoV-2 and related sarbecoviruses, thereby addressing the pressing need for a more universally protective vaccine.
Experimental Process
- Antigen Design
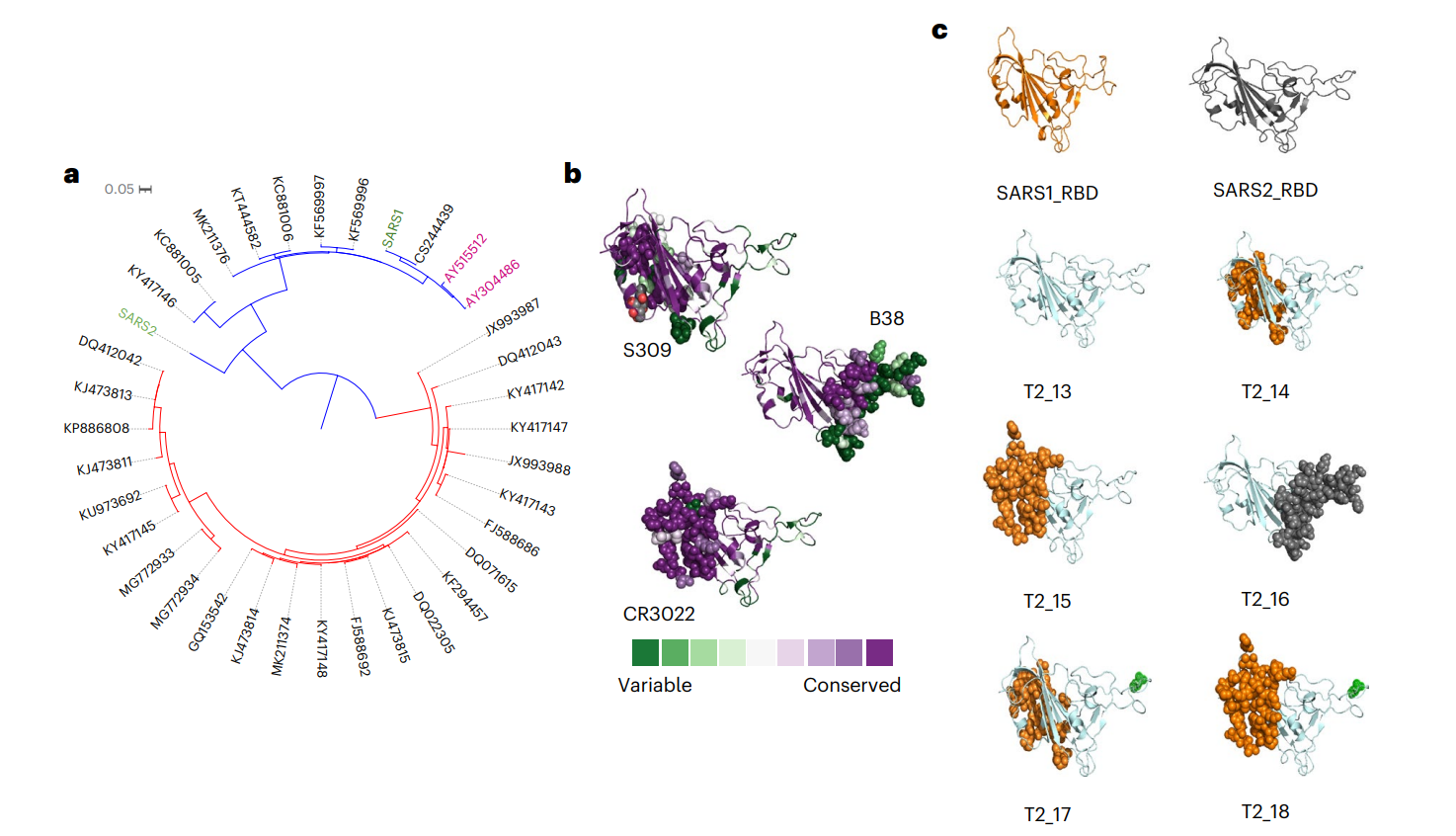
Figure 1. In silico design of antigen candidates
- Key Steps:
The research team utilized a viral-genome-informed computational approach to select and engineer immune-optimized antigens based on the receptor binding domain (RBD) of the spike proteins from various sarbecoviruses. A phylogenetic tree was constructed to identify conserved and distinct epitopes across different strains. - Results and Key Data:
The optimized antigen sequence was designated as T2_17, which was structurally modeled and analyzed for stability using the FoldX algorithm. The antigen showed a high degree of conservation across multiple strains of sarbecoviruses. - Significance of the Result:
By focusing on conserved regions, the antigen was expected to elicit broad immune responses against multiple strains, an essential feature for a vaccine aimed at preventing zoonotic spillovers. - Key Innovations:
The use of a computational design approach allowed for the rapid identification and synthesis of an antigen that could potentially target a wide range of sarbecoviruses, overcoming limitations of traditional vaccine development methods.
- In Vivo Immunization in BALB/c Mice
- Key Steps: BALB/c mice were immunized with the designed antigens, including the T2_17 and SARS-CoV-2 RBD, delivered as DNA immunogens. Blood samples were collected at 15-day intervals for serological analysis.
- Results and Key Data: Serum analyses demonstrated significant cross-reactive antibodies against spike proteins from SARS-CoV, SARS-CoV-2, WIV16, and RaTG13. Mice immunized with T2_17 showed comparable binding profiles to those vaccinated with SARS-CoV-2 RBD, indicating effective immune response.
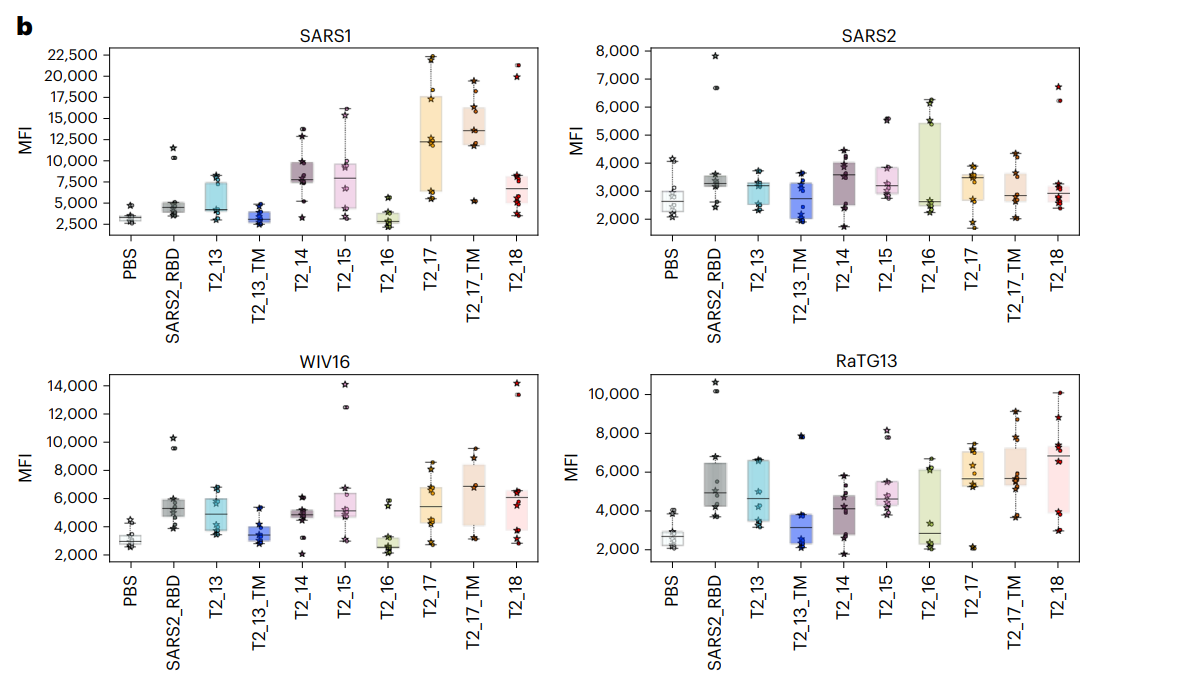
Figure 2. In vivo immunogenicity of antigens in BALB/c mice
- Significance of the Result: The results confirmed that the T2_17 antigen successfully induced an immune response similar to that of a well-established antigen, suggesting its potential as an effective vaccine candidate.
- Key Innovations: The incorporation of multiple spike variants in the immunogenicity assessment provided a comprehensive evaluation of the antigen’s effectiveness, highlighting its broad applicability.
- Cross-Reactivity Assessment
- Key Steps: Sera from immunized mice were subjected to flow cytometry and enzyme-linked immunosorbent assays (ELISA) to evaluate binding and neutralizing antibody responses against various spike proteins.
- Results and Key Data: The analysis revealed that T2_17 elicited binding antibodies that significantly neutralized pseudoviruses expressing SARS-CoV, SARS-CoV-2, WIV16, and RaTG13. Notably, T2_17 showed higher neutralization efficacy against RaTG13 and SARS-CoV compared to other control antigens.
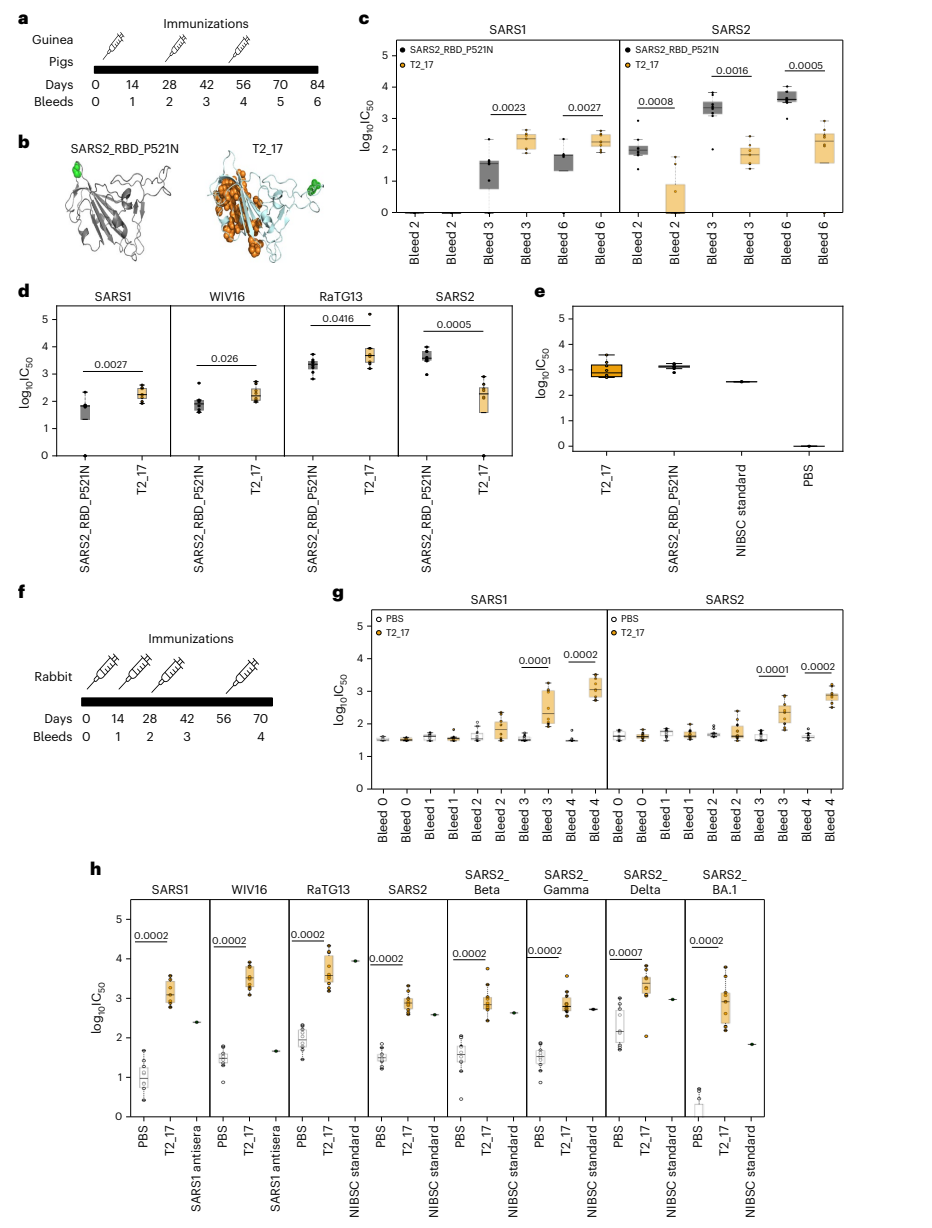
Figure 3. Immunogenicity studies in guinea pigs and rabbits
- Significance of the Result:
These findings illustrated the potential of T2_17 to provide cross-protection against diverse strains, which is crucial for addressing the evolving nature of coronaviruses. - Key Innovations: The study employed pseudovirus neutralization assays, allowing for a more accurate assessment of the functional capacity of elicited antibodies in a laboratory setting.
- Immunogenicity Confirmation in Outbred Guinea Pigs
- Key Steps: Outbred guinea pigs were immunized using a needleless delivery device to ensure consistent intradermal administration of T2_17. Blood samples were obtained periodically to monitor antibody responses.
- Results and Key Data: The serum analysis indicated that T2_17 induced robust binding antibodies and neutralizing responses against both SARS-CoV and SARS-CoV-2 after multiple immunizations. Notably, T2_17 exhibited superior neutralizing antibody levels against SARS-CoV compared to a control antigen (SARS2_RBD_P521N).
- Significance of the Result: This confirmed the antigen’s potential for broad immunogenicity in a more genetically diverse population, which is essential for vaccine development.
- Key Innovations: The study utilized a standardized needleless immunization approach, improving the consistency and safety of vaccine delivery.
- Challenge Studies in K18-hACE-2 Mice
- Key Steps: K18-hACE-2 transgenic mice were initially primed with the AZD1222 vaccine and subsequently boosted with T2_17, followed by a challenge with either the Victoria strain or the Delta variant of SARS-CoV-2.
- Results and Key Data: Mice that received the T2_17 boost demonstrated significantly higher neutralizing antibody titers against the Delta variant compared to those boosted with AZD1222 alone. Importantly, all immunized groups survived the viral challenge.
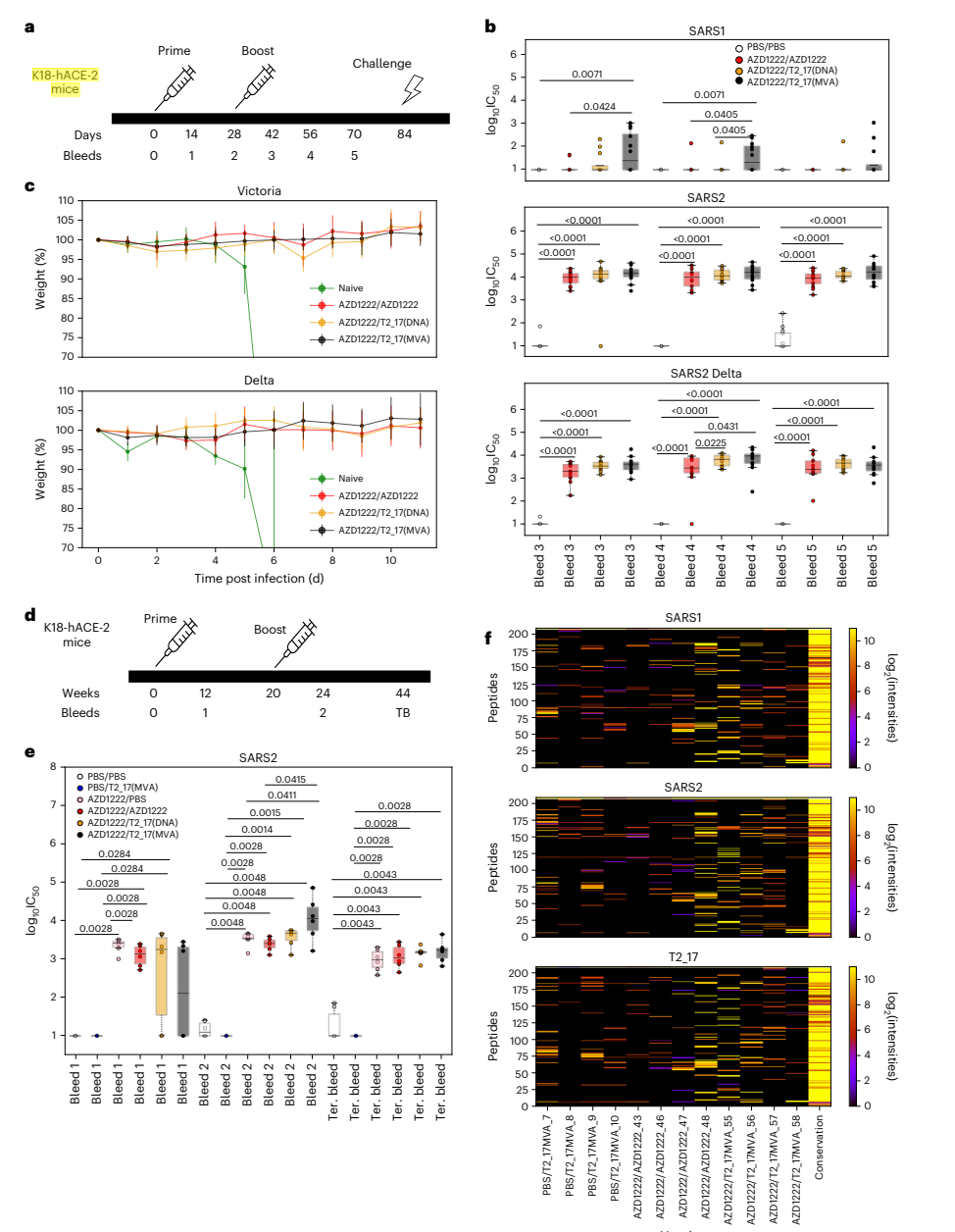
Figure 4. Immunogenicity and challenge studies in K18-hACE-2 mice.
- Significance of the Result: This demonstrates the potential of T2_17 to enhance immune responses in individuals with prior vaccination, reinforcing its role as a promising booster candidate.
- Key Innovations: The combination of a heterologous prime-boost strategy using T2_17 provided a novel approach to improving vaccine efficacy against variants of concern.
- Messenger RNA (mRNA) Vaccine Validation
- Key Steps: To assess the mRNA delivery system, T2_17 was formulated as an mRNA vaccine and administered to BALB/c mice. The mice were assessed for immune responses following administration.
- Results and Key Data: The mRNA-based T2_17 generated significant binding and neutralizing antibodies against SARS-CoV-2, with the membrane-anchored version (T2_17_TM) showing enhanced responses compared to the soluble form.
- Significance of the Result: This highlighted the versatility of T2_17 as a vaccine candidate across multiple delivery platforms, including mRNA technology, and its potential for rapid adaptation to emerging variants.
- Key Innovations: The development of a membrane-anchored mRNA vaccine demonstrated an innovative approach to enhancing immunogenicity and neutralization capacity, potentially leading to improved vaccine formulations in the future.
Conclusion
The findings from this study indicate that the computationally designed antigen T2_17 can elicit broad immune responses, providing a promising avenue for the development of vaccines against SARS-CoV-2 and its variants. The results suggest that such a vaccine could play a crucial role in controlling future outbreaks and preventing zoonotic spillovers.
However, while the study demonstrates significant potential, it also acknowledges limitations related to the scope of animal models used and the need for further validation in human clinical trials. Future research should focus on optimizing the antigen further and assessing its effectiveness against newer variants that continue to emerge globally.
As the pandemic evolves, the development of versatile vaccines such as T2_17 could be pivotal in achieving long-term control of coronaviruses and safeguarding public health.
Reference
Vishwanath, Sneha, et al. “A computationally designed antigen eliciting broad humoral responses against SARS-CoV-2 and related sarbecoviruses.” Nature Biomedical Engineering (2023): 1-14.
